R. Beltrami, D.
Medini, C. Donati, N. Pacchiani, A. Muzzi, A. Covacci
IRIS
Bioinformatic Unit, Chiron S.p.A.
biocomp_admin_siena@chiron.it
1.
Introduction
2.
Workpackage activity summary
3.
Biowulf system setup
4.
Blast reconstruction and recompilation
5.
Gromacs reconstruction and recompilation
6.
Future developments
7.
Appendix A
8.
Appendix B
9.
Appendix C
1. Introduction
Top
According to the objectives of this workpackage, the
hardware architecture and the software solution developed on the
prototype system have been customized and deployed on the production
system in order to be optimized and tuned and finally used for the
company's genomics projects.
Two basic software tools have
been taken into account: NCBI's blast toolkit and GROMACS. During the
workpackage 3[i] both have been adapted and customized in order to be
used on the Biowulf cluster and significant biological sequence
databases have been compiled to perform test and optimization on the
production system. A Symmetrical Multi Processor system has been used
during the test phase not only as a comparison machine but also to
verify the usability of the software tools on more traditional
architecture as well.
Finally, future activities will be
outlined and the tuning and optimization strategies will be
described. These tasks will be the objectives of the final
workpackage 5.
2 Workpackage activity summary
Top
Workpackage 4 is composed of two main phases: i)
construction of the production system and deployment of the software
tools; ii) design and compilation of production databank.
Phase
I.A) Firstly, the hardware acquisition and setting has been done[ii].
Three Sun Microsystems SunBlade 1000 have been purchased and the
operating system configuration completed in order to be part of the
Biowulf cluster. Also the other nodes have been configured.
Phase
I.B) The software developed and used in the prototype system have
been configured, compiled and installed on the production system. All
the additional system configurations have been made and the required
software libraries and tools have been installed in order to run the
selected tools. More specifically the Python language
interpreter[iii] (version 2.1.1), the mxTools package[iv] and the Sun
High Performance Computing ClusterTools[v] (version 4.0) have been
installed. NCBI's blast[vi] and the DNC blast[vii] have been compiled
and GROMACS suite[viii] has been configured to use the Sun MPI
implementation and then compiled. All the compilation jobs have been
done using the Sun Forte C compiler version 6 update 2, fully 64-bit
compliant. A number of query sequences and test databanks have also
been defined and built.
Phase II) Finally, the production
databanks have been built. We have selected two public available
databank, the NCBI GenBank[ix] (release 124.0, June 2001) and the Non
Redundant Protein Databank[x] (June 2001), and two custom made
databanks (one for DNA and one for amino-acidic sequences): 36 public
plus 5 private completed bacteria genome sequences. Given the
structure of those databanks a naïve splitting of the databanks
among the cluster node could result in biased searches where most of
the nodes complete the run in far less time than the others. Thus a
databank "randomization" have been developed and applied to
all the databanks.
During the workpackage 4 some preliminary
tests have been done[xi]. According to the results obtained some
optimization and tuning steps have been identified to improve the
overall performances and the limit of this solution outlined.
3
Biowulf system setup
Top
The available Network of Workstation is made up of 10
nodes with up to 21 CPUs[xii] connected through a switched
Fast/Gigabit-Ethernet. Reflecting the on-site logistic distribution
of the hardware resources, some of the nodes are connected through
different Fast-Ethernet switches, all along a Gigabit backbone. All
the nodes run Sun Solaris operating system from release 2.6 up to
8.
We have defined a biowulf computational user on all the
nodes of the Network, sharing the nfs-exported home directory from
the front-end node. To speed up the execution of remote shell (rsh)
commands through the nodes, the home directory has been statically
nfs-mounted on all the nodes, leaving the use of the Solaris daemon
automounter to the standard users. We have chosen the C shell csh as
the default shell in order to facilitate the automatic initialization
of a set of GROMACS[xiii] setup scripts. A local /opt/biowulf
directory tree has been used on all the nodes to store local data
(local databanks volumes[xiv], MPI local resources and GROMACS
dynamically linked shared libraries). All the nodes have been
reciprocally trusted, so that the biowulf user can execute local or
rsh commands transparently.
We have compiled all the software
with the 64-bit FORTE Sun C, C++, FORTRAN Compiler suite, with
architecture-specific optimization. We have found a performance
increase of 20% to 40% on most routines with respect to the 32- bit
GNU gcc compiled versions.
The DNC implementation[xv] of
blast[xvi] makes direct use of the UNIX socket to establish the
inter-node communication, thus no extra message passing tool is
needed. On the front-end node the Python interpreter version 2.1.1
has been 64-bit compiled and installed, together with the mxTools
package add-on, available under the e.Genix Public License Agreement.
In this first implementation we have used an heterogeneous subset of
the available nodes, namely: three Sunblade-1000 (2 UltraSparc III
750 MHz CPUs, 2 GB RAM, 2x36 GB UltraSCSI FC-AL HD, Solaris 8), one
Ultra-60 (2 UltraSparc II 360 MHz CPUs, 1 G RAM, 2x9 GB UltraSCSI HD,
Solaris 2.7) and one Ultra-10 (1 UltraSparc II 360 MHz CPU, 512 MB
RAM, 2x9 GB UltraSCSI HD, Solaris 8), for a total of 5 nodes and 9
CPUs in cluster. One of the Sunblade-1000 has been employed both as a
computational node and as the front-end of the system. These
workstations can be used for other tasks and not only dedicated to
the cluster. However off-peak hours the cluster is planned to be
employed as "blast engine", with no need of other specific
system tuning, thus exploiting the available computing power more
completely. For comparison a Symmetric Multi Processor server (Sun
Enterprise 4500, 8 UltraSparc II 400 MHz CPUs, 8 GB RAM, 10x36 GB
UltraSCSI HD, Solaris operating system release 2.7.) has also been
used.
The parallel version of the GROMACS molecular dynamics
engine (mdrun) requires either a Parallel Virtual Machine (PVM[xvii])
or a Message Passing Interface (MPI[xviii]) communication layer. We
have tested two MPI implementations, both fully MPI-1.2 and partially
MPI-2 compliant: MPICH[xix] version 1.2.2, according to the prototype
implementation[xx], and the SUN MPI, which is a part of the SUN High
Performance Computing ClusterTool[xxi] version 4.0. Both are freely
available and well documented, the former under a specific Government
License, the latter under the Sun Community Source Licensing program.
For this first implementation a high performance symmetric subset of
the available nodes has been used, composed of the three
Sunblade-1000 (2 UltraSparc III 750 MHz CPUs, 2 GB RAM, 2x36 GB
UltraSCSI FC-AL HD, Solaris 8) for a total of 3 nodes and 6 CPUs in
cluster.
4 Blast reconstruction and recompilation
Top
NCBI's blast program has become the reference standard tools
for biological sequence homology search. Since its first release
blast has undergone several improvements and many important changes
were included especially in the version 2.0; we used the latest
release dated April 5, 2001 included in the NCBI toolkit. We have
compiled the whole suite in a fully 64 bit environment with the SUN
Forte C compiler, version 6 update 2. For the homology search
routines (namely blastp, blastn, blastx, tblastn, tblastx) a "-xO3
-xarch=v9a" optimization has been adopted. It has shown to be
the optimal choice (in terms of single and double processor
performance) with respect to other SUN Forte optimization, (such as
the complex -fast option) being as twice as faster than a
gcc-compiled 32-bit version.
We started with two publicly
available databanks such as the NCBI's GenBank release 124.0 (June
2001) and the Non Redundant protein DB (June 2001). We have used the
indexing routine formatdb of the NCBI toolkit to translate the
multi-FASTA formatted databanks into the blast-readable format. The
overall dimension of the indexed databanks is shown in Table 1.
|Non Redundant Protein DB | |(NR): | 705002 entries | 334.5 MB | |Genbank: |1429009 entries |1454.6 MB |
Table 1
The
formatdb multi-volume option has also been adopted in order to obtain
multi-volume (2, 4, 6, 8 and 9-volumes) versions of the same
databanks to be used with the distributed DNC blast. There is no
relevant difference in the overall dimension of the databanks between
the single and the multi- volume versions. We have stored the
complete versions of the databanks on the SMP server and on one of
the cluster nodes (one of the Sunblade-1000) in their respective
local /opt/biowulf directories; then we have distributed some
databank volumes to all the nodes in the cluster on their respective
local storage systems as well. All the multi-volume versions have
also been put onto the local disks of the SMP for comparison
purposes.
Finally, we have selected fourteen (seven DNA and
seven protein) heterogeneous input queries: for each set we have
three bacterial, two human and two yeast sequences. The detailed
composition and character length of the query set is reported in
Appendix A.
We have performed blastp and blastn alignments of
the fourteen sequences against the GenBank and the NonRedundant DB
using the standard NCBI routines (enabling the multithread support up
to 8 CPUs) on the SMP server and using the DNC implementation both on
the cluster (using 1, 2, 4, 6, 8 and 9 CPUs) and on the SMP server.
We have obtained identical blast reports in all the three conditions,
apart from the negligible numerical discrepancy in the E-value[xxii]
already discussed in the prototype implementation. Detailed
quantitative results of these tests are reported
elsewhere[xxiii].
5 Gromacs reconstruction and
recompilation Top
GROMACS is a versatile computational chemistry and molecular
modeling package, containing a Molecular Dynamics engine, a number of
pre-MD tools needed for the simulation setup, and several post-MD
data analysis tools, for a total of about 65 binaries. The MD engine
program (mdrun) is actually a parser for a set of different
simulation integrators (energy minimization, molecular dynamics,
essential dynamics, umbrella sampling, stochastic dynamics, potential
of mean force, simulated annealing) and is the only parallel portion
of the suite.
The compilation of the suite is made easy
through a hierarchical distribution of Makefiles. General
compile-time options can be set in the uppermost one, and a set of
pre-defined Makefiles for different platforms is provided with the
software. At a first stage we have adopted the standard Ultra-SPARC
single-precision version provided with the source tree with only
minor changes, and we have compiled and installed MPICH on the
cluster nodes. MPICH adopts the concept of device to identify the
communication transport medium. In our conditions two devices can be
adopted: the ch_p4 device, designed for the TCP/IP transport over
network connection, and the ch_shmem device for communication over
SMP architectures (to avoid the TCP/IP stack delay in communications
among the two CPUs on the same host). We just had to provide the
correct propagation of the environment variables from the master
shell interpreter to the remote ones running the slave processes to
let the compiled software run right out of the box. Unfortunately, as
it was expected, the first performance tests showed the same bad
scaling behavior [pic] as in the prototype testing[xxiv], the only
difference being the absence of the aggregate-cache effect at low CPU
number present therein, due to the 8 MB internal cache of each CPU in
the cluster.
The low quality of the network transport has been
identified as one of the key problems. In order to improve the
communication performance without increasing the Total Costs of
Ownership we have tried to tune the communication software. Rather
than one of the several portable MPI implementations available
(mpich, lam, mpilite among the others) we have chosen a
Solaris-platform specific one, the SUN-MPI, because of it's high
integration with the SUN High Performance Computing ClusterTool that
includes the parallel Sun Scalable Scientific Subroutine Library (Sun
S3L). This allowed us to take advantage of the high parallel
optimization capability of the 64-bit FORTE SUN compiler. We have
modified the standard Makefile with specific sparcV9 optimization,
linking the high performance mpi and s3l libraries, allowing for
shared-libraries use, forcing the alignment of common block data; the
resulting Makefile is included in Appendix B.
Moreover, we
have modified the csh initialization script that the GROMACS suite
uses to set the required environment variables setting and some MPI-
specific option. Namely, we have suppressed cyclic message passing
because we don't need any extra synchronization between sender and
receiver, suppressed general polling because the code uses system
buffers safely, increased the MPI consumption of shared memory thanks
to the 1 GB RAM memory per CPU available. The customized GMXRC
initialization file is also included in Appendix B.
Finally,
since light-load latent processes (such as standard UNIX daemons) and
external network traffic can have a relevant impact on the
performances of such an intensive software, we have characterized a
minimal set of system processes to remain active when the node is
used in parallel processing and we have connected the three nodes
with a dedicated.
Under these conditions we have performed a
short molecular dynamics simulation on three sample systems: a small
peptide in water (Small_Pep), a medium sized di-proteic system in
water (Hiv_prot) and a big Betaglycosidase also in water (?-Glyc).
Table 2 contains a summary of the three system specifications.
By
running each simulation at 1 and 2 CPUs on the same node it is
possible to obtain, for each system, a rough estimate of the fraction
phi of parallel code in the well known Amdahl's law:
S(n,phi) = [(1-phi) + phi/n]-1
where S is the speedup as a function of phi and of the CPU
number n. The phi values are also listed in Table 2.
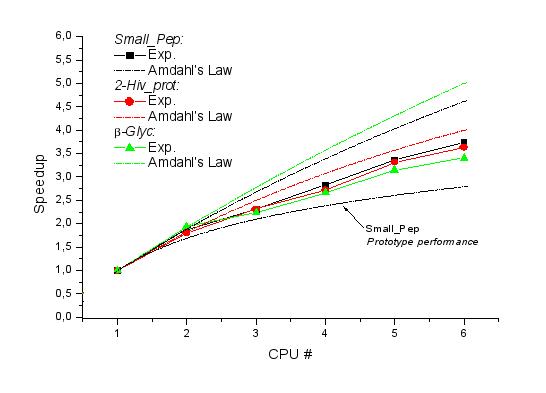
Then the
simulations have been executed on 3, 4, 5 and 6 CPUs; the results are
shown in Fig. 1 together with the estimated Amdahl's laws and the
prototype performance reference.

Fig. 1
|System |Residue|Prot. |Water |Total Atom|Long-Ran|? |Simulation | | |s # |Atoms |molecule|# |ge | |length | | | |# |s # | |Electros| | | | | | | | |tatic | | | |Small_P|34 |270 |2794 |8655 |ON |0.94|10000 steps | |ep | | | | | | |(20 ps) | |Hiv_pro|99 + |952 + |5893 |19584 |ON |0.90|5000 steps | |t |99 |953 | | | | |(5 ps) | |?-Glyc |489 |5249 |24083 |77505 |ON |0.97|5000 steps | | | | | | | | |(10 ps) |
Table 2
The decrease in the
speedup at 3 CPUs due to the presence of inter-node communications is
clearly evident. Also, the biggest system (i.e. the more
computationally challenging and more relevant for real-case
applications) is clearly the more highly affected by the standard
commercial network employed. Nevertheless, we can appreciate at this
first stage a marked increase in the scalability with respect to the
prototype testing case, going from the 46% at 6 CPUs for the
Small_Pep system to the 63%, for the same system, in the present
case.
Database selection and indexing
The
continuously growing amount of available genetic information provides
larger and larger sources of information, with the obvious advantage
of an increasingly general reference for similarity searches, but
also with the obvious disadvantage of higher search times. As a
consequence, we have decided to build a database made of four
components: two, publicly available and widely used collections of
genes (GenBank release 124.0, June 2001) and expressed proteins
(NonRedundant database, June 2001), and two custom collections of
specific bacterial genomes Open Reading Frames (named Bacteria_N) and
the corresponding expressed proteins (Bacteria_P). The latter are
partially publicly available, partially private proprietary data. The
GenBank is a huge standard reference for nucleotide sequences while
the NonRedundant database is for proteins. The NonRedundant has the
advantage of being far smaller thanks to the merging of different
annotations of the same protein sequence into a single one. These
huge collections of data are essential references in any genomic
research, but also specific subsections can be crucial in focusing on
particular research targets. In fact custom DBs are commonly used in
industrial production environments where proprietary genomic
sequencing projects are used.
Henceforth we have collected in
Bacteria_N and Bacteria_P all the Open Reading Frames of the
following 41 full sequenced bacterial genomes (36 publicly available
and 5 proprietary data) with the respective expressed proteins:
1:
Aeropyrum_pernix_K1
2: Aquifex_aeolicus
3:
Archeoglobus_fulgidus
4: Bacillus_halodurans
5:
Bacillus_subtilis
6: Borrelia_burgdoferi
7: Buchnera_sp.APS
8:
Campylobacter_jejuni
9: Chlamydia_muridarum
10:
Chlamydia_trachomatis
11: Chlamydiophila_pneumoniae_AR39
12:
Chlamydiophila_pneumoniae_J138
13: Deinococcus_radiodurans
14:
Escherichia_coliK12
15: Haemophylus_influenzae
16:
Halobacterium_sp.NRC-1
17: Helicobacter_pilori_26695
18:
Helicobacter_pylori_J99
19:
Methanobacterium_thermoautotrophicum_deltaH
20:
Methanococcus_jannaschii
21: Mycobacterium_tubercolosis_H37Rv
22:
Mycoplasma_genitalium
23: Mycoplasma_pneumoniae
24:
Neisseria_meningitidis_MC58
25: Neisseria_meningitidis_Z2491
26:
Pseudomonas_aeruginosa_PA01
27: Pyrococcus_abyssi
28:
Pyrococcus_hirikoshii_OT3
29: Rickettsia_prowazekii_MadridE
30:
Synechocystis_PC6803
31: Thermoplasma_acidophilum
32:
Thermotoga_maritima
33: Treponema_pallidum
34:
Ureaplasma_urealyticum
35: Vibrio_cholerae
36:
Xylella_fastidiosa
37 - 41: five complete bacterial genomes based
on private corporate data.
Then we have indexed the four
multi-FASTA databanks, by means of the NCBI toolkit's indexing
routine formatdb, in several multi-volume versions. The dimensions of
the four databanks in the indexed versions is reported in Table
1.
From the results of a set of preliminary test performed on
the Biowulf cluster[?] we have decided to maintain two versions of
the same databanks on the cluster node local storages: a 6-volumes
version, stored on the 3 fast dual-processor SunBlade 1000, and a
9-volumes version to be shared among all the nodes in cluster. The
first data set, at the present stage of development of the DNC-Blast
software[?], is the preferred one if the mentioned faster nodes are
free from other computational load. If some of them are concurrently
used for other tasks, the second choice is more likely to give better
performance.
|Non Redundant Protein database (NR): |334.5 Mb | |Bacteria Open Reading Frames (Bacteria_N): |21.6 Mb | |Bacteria Proteins (Bacteria_P): |28.9 Mb | |Genbank: |1454.6 Mb |
Table 3
Database
randomization
The mentioned preliminary tests showed
that, in order to increase the intrinsic load-balance in homogeneous
multi-volume blast searches, the first requirement is the
randomization of the searched databank. In fact, when a query
sequence is searched against a portion (volume) of the databank, the
execution-time is almost proportional to the number of hits with the
database entries. In the original database the sequences are usually
organized in blocks of different organisms, so there are very good
chances to have a highly non-homogeneous number of hits among the
different volumes. To avoid this, we have elaborated a randomizing
procedure that "scrambles" the sequences in a generic
multi-FASTA database, then the usual multi-volume indexing can be
performed to obtain a set of blast- readable database files.
The
Perl program, named dbrandom, that implements this procedure is
included in Appendix C. It simply reads the original multi-FASTA
file, assigns a random generated number as a label to each sequence,
and prints the randomized database sorted on the label itself. The
dimension of the original and of the randomized file is identical. We
have applied this procedure to all the databanks indicated in the
previous section and re- indexed in the multi-volume versions all of
them. This set of files, shared among the Biowulf nodes and on the
SMP server according to the above mentioned procedure, constitutes
the production database.
6 Future developments Top
As far as the implementation of the DNC blast onto the
Biowulf architecture in concerned, some considerations are mandatory.
The asymmetry of the system is a key feature of the system: it
exploits a set of computational resources already available on-site
but only partially (or non- continuously) utilized. In this
connection, the main limitation of this first version of the DNC
blast is that it submits all the sub-jobs at once. In order to avoid
the slow-down of FPU and I/O-jam produced by many sub- jobs sent
concurrently to the same processor, one is forced to create exactly n
sub-jobs for n CPUs available on the cluster. The net effect is that
the slowest CPU determines the overall speed of the process. However,
this problem can easily be worked out: for each databank the
"optimal" (and fixed) number m of slices will be a small
value for the short databanks while might be much greater than the
number n of the available CPUs for the larger databanks. In the
latter case, a DNC blast Phase II is needed in order to have a blast
query divided into m sub-processes. Only the first n are initially
submitted to nodes and the remaining ones are sequentially queued as
the first ones are done. In this way faster processors should execute
more sub-jobs than the slower ones thus balancing the load on the
nodes. This phase is already under development, and results will be
reported in the deliverable D 5.1.
Moreover, a DNC blast Phase
III should also be developed, in order to take into account for
concurrent usage of the cluster by several users (i.e. query queuing
issues) and possible lack of total RAM in the cluster with respect to
the global dimension of the databanks used (thus the I/O cache
effect[?] is diminished). This last phase should consist in the
set-up of a "smart queuing and scheduling system" aware of
the current state of the databanks in the cluster in order to choose
the most appropriate sub-set of nodes to be assigned the jobs in
queue. This queuing system shall take into account for the speed of
specific nodes, location and cache state of the slices to search
against and assign the queued jobs in order to minimize the databank
slides reloads. Such a system appears to be mandatory in order to
have the Biowulf cluster actually working as a DNC blast server in a
real production environment. This phase will be developed in the WP5
and results will be reported in the deliverable D5.3.
Also the
tuning of Gromacs on the Biowulf platform is already under study.
Here we have to face two kinds of problems: the network communication
performances and the specific use of the nodes for the computational
tasks. The inter-node communication can be schematically divided into
three components: the MPI layer, the TCP/IP layer and the network
hardware. In the vein of the present project objectives, we are
trying to obtain better performances from the commodity hardware
itself, without increasing the total cost of the cluster because of
the acquisition of dedicated high- price network hardware. We have
already shown that acting on the MPI layer we can achieve a good
improvement on the communication performance. The second step is the
TCP/IP layer. The implementation of this protocol is typically
optimized for insecure, Wide Area Network communications, and
specific tuning of the TCP/IP parameters for a switched Local Area
Network environment, while slightly affecting the network
performances on Internet applications, could remarkably help in the
case of study. Also, the option of switching from TCP/IP to UDP/IP as
the transport layer for MPI could be carefully considered.
Finally,
we have to observe that for computational tasks such as Molecular
Dynamics (MD) simulations, we cannot use the Cluster nodes
promiscuously, i.e. the hosts must be as free as possible from any
kind of load, and network connection must be strictly reserved to the
simulations. To this end, we are studying the option of defining a
dedicated Unix runlevel that defines all the required system
settings, so that all the machine configurations for MD jobs are
reduced to an appropriate init call.
7 Appendix A
Top
Query sequences selection:
DNA sequences
|Sequence |String length | | | | |>gb|AE000511|AE000511:628313-629785, HP0595 |1472 char. | |>gb|AE005672|AE005672:11812-12354 |542 char. | |>gb|AE004091|AE004091:c4060591-4059956 |634 char. | |>AB000475 AB000475 Yeast mRNA for S.pombe TFA2 |1006 char. | |homolog, partial cds. 2/1999 | | |>AB000518 AB000518 Fission yeast mRNA for |939 chars. | |translation initiation factor eIF1A, partial cds. | | |2/1999 | | |>gi|15297278:406241-520784 partial |615 char. | |>gi|15296557:3421230-3438809 partial |1055 char. | |Protein sequences | |Sequence |String length | |>gi|7226763|gb|AAF41875.1| thiol:disulfide |513 char. | |interchange protein DsbD | | |>gi|9949490|gb|AAG06744.1|AE004758_1 conserved |1279 char. | |hypothetical protein [Pseudomonas aeruginosa] | | |>gi|14971470|gb|AAK74199.1| transcription-repair |781 char. | |coupling factor [Streptococcus pneumoniae] | | |>gi|1003002|gb|AAA77064.1| (U34887) beta-lactamase|364 char. | |[Cloning vector pRSQ] | | |>gi|2506773|sp|P04173|LEU3_YEAST 3-ISOPROPYLMALATE|678 char. | |DEHYDROGENASE | | |>gi|13637515|ref|XP_002644.2| ATP-binding |829 char. | |cassette, sub-family B (MDR/TAP), member 11 [Homo | | |sapiens] | | |>gi|13636078|ref|XP_017752.1| hypothetical protein|371 char. | |FLJ22555 [Homo sapiens] | |
8 Appendix B
Top
Gromacs custom Makefile:
# Sun Ultrasparc
#
# GROMACS - Groningen Machine for Chemical Simulation
# Copyright (c) 1990, 1991, 1992, Groningen University
#
# Makefile for gromacs on ultrsparc processor
#
# Duccio Medini, modified 09/20/2001 for
# SUN HPC ClusterTools MPI
#
#
SYSDEFS = -DHAVE_IDENT -DHAVE_STRDUP -DHAVE_STRCASECMP -DFINVSQRT
-DCINVSQRT
# Sun workshop compilers
CC = mpcc
CCC = mpCC
F77 = mpf90
# This is intended for ultrasparc architecture, and allows for
semidangerous optimization
#
CFLAGS = -KPIC -xO3 -fast -xarch=native64 -lmpi -lmopt -ls3l
CCFLAGS = -KPIC -xO3 -fast -xarch=native64 -lmpi -lmopt -ls3l
FFLAGS = -dalign -KPIC -xO3 -fast -xarch=native64 -lmpi -lmopt -ls3l
# Generic linking stuff
LDFLAGS = -L$(LIBDIR) -L/usr/lib -L/usr/openwin/lib \
-L/opt/biowulf/SUNWhpc/HPC4.0/lib/sparcv9 \
-xO3 -fast -xarch=native64 -lmpi -lmopt -ls3l
LD = $(F77) $(LDFLAGS) -z nodefs
FLD = $(F77) $(LDFLAGS)
CCLD = $(CCC) $(LDFLAGS) -z nodefs
XLIBS = -lsocket -lX11
SYSLIBS = $(LINKLIB) -lmopt -lnsl
SHAREIT = (cd $(LIBDIR); ar x $(LIB); cc $(LDFLAGS) \
-o $(SONAME) -G *.o; $(RM) *.o)
RANLIB = echo
X11INC = -I/usr/openwin/include
#
# USER MODIFIABLE SECTION
#
# If you want to use fortran innerloops set this to yes
# For most machines this will improve the performance quite a bit
# because C-compilers are not as good as Fortran compilers
USEF77 = yes
SYSDEFS += -DF77UNDERSCORE
#
# If you want to run in *** P A R A L L E L ***
# please select either PVM or MPI.
#
USE_PVM3 = no
USE_MPI = yes
#
# If you want to use compressed data files set this to yes
# This uses the xdr libraries of your UNIX system, which is virtually
# always present, because it is necessary for NFS (network file system)
#
USE_XDR = yes
#
# Graphics Library
# Set this on if you have Open GL and the Viewkit library
# (standard with SGI, available for money elsewhere)
# This is used in the Open GL trajectory viewer.
HAVE_GL = yes
#
# End
Gromacs GMXRC initialization script:
#! /bin/csh
# Directories, edit to match your site, we assume
# users have a c-shell always
#
# Duccio Medini, Modified 09/12/2001
# for use with the SUN MPI.
# remove previous GROMACS environment, if present
source /opt/biowulf/gmx2.0/NOGMX -quiet
# This is were the sources are
setenv GMXHOME /opt/biowulf/gmx2.0
#
# If you support multiple machines then it's useful to
# have a switch statement here, which, depending on hostname
# point to the proper directories, and sets the GMXCPU variable
#
# For easy updating, it is also recommended to put you local
# stuff in the GMXRC.local file
# Don't forget the GMXCPU variable
#
setenv LOCAL_RC $GMXHOME/GMXRC.local
if ( -f $LOCAL_RC ) then
source $LOCAL_RC
else
# Ultra SPARC definition
setenv GMXCPU ult
endif
# Some directories that live below the root
setenv GMXBIN $GMXHOME/bin/$GMXCPU
setenv GMXLIB $GMXHOME/top
#
# Default Graphics Font
#
setenv GMXFONT 10x20
#
# Set the path #
setenv PATH "$PATH":"$GMXBIN"
#
if ( $?LD_LIBRARY_PATH ) then
setenv LD_LIBRARY_PATH "$LD_LIBRARY_PATH":$GMXHOME/lib/$GMXCPU
else
setenv LD_LIBRARY_PATH $GMXHOME/lib/$GMXCPU
endif
if ( $?MANPATH ) then
setenv MANPATH "$MANPATH":$GMXHOME/man
else
setenv MANPATH /usr/man:$GMXHOME/man
endif
#
# Specific SUN MPI settings
#
setenv MPI_SHM_CYCLESTART 0x7fffffffffffffff
setenv MPI_POLLALL 0
setenv MPI_SHM_SBPOOLSIZE 8000000
setenv MPI_SHM_NUMPOSTBOX 256
# end of script
9 Appendix C
Top
dbrandom.pl :
#!/usr/local/bin/perl
#
# Databank Randomizer: reads a MULTI-FASTA formatted
# sequence database and writes the randomised database
# in the same format.
#
# USAGE: dbrandom.pl input_file output_file record_number
#
# where record_number is the number of entries
# in the database.
#
# Claudio Donati, October 3 2001.
#
# COPYRIGHT NOTICE:
#
# The following is proprietary code of Chiron S.p.A.
# Any use of the whole code or a part of it has to
# be explicitly approved by the authors.
# For more information contact biocomp_admin_siena@chiron.it
#
open INPUT, $ARGV[0] ;
open TEMP1, ">temp1" ;
while($line=< INPUT >)
{
if($line=~">")
{
print TEMP1 "\n";
$numero=1000*$ARGV[2]*rand;
$numero=int $numero;
chop $line;
print TEMP1 "$numero\{\}$line\{\}";
}
else
{
chop $line;
print TEMP1 "$line";
}
}
close TEMP1 ;
close INPUT ;
system "sort -n -t { ./temp1 -o ./temp2";
exec "rm -f ./temp1" ;
open TEMP2, "temp2" ;
open OUTPUT, ">$ARGV[1]" ;
while ($line=)
{
chop $line;
$indi=0;
@a=split /{}/,$line;
print OUTPUT "$a[1]\n";
$lun=length $a[2];
while ($indi<$lun)
{
$sotto=substr $a[2],$indi,80;
print OUTPUT "$sotto\n";
$indi+=80;
}
}
close TEMP2 ;
close OUTPUT ;
exec "rm -f ./temp2" ;
# END
- IRIS - Chiron S.p.A., Via Fiorentina 1,
53100 Siena. ITALY.
-----------------------
[i] Rossi, I.
and Fariselli, P. Deliverable 3.1 of the present Project; Rossi, I.
and Martelli, P.L. Deliverable 3.3 of the present Project.
[ii]
Beltrami, R. et al., Deliverable 4.1 of the present Project.
[iii]
See http://www.python.org
[iv] See
http://www.lemburg.com/files/python/mxTools.html
[v] See
http://www.sun.com/software/hpc
[vi] Downloaded as a part of the
NCBI Toolkit from ftp://ftp.ncbi.nlm.nih.gov
[vii] Rossi, I. and
Fariselli, P. cited.
[viii] Berendsen, H.J.C., van der Spoel, D.
and van Drunen, R., GROMACS: A message-passing parallel molecular
dynamics implementation, Comp. Phys. Comm. 91 (1995) 43-56.
[ix]
Wheeler, D.L. et al., Nucleic Acids Research 29 (2001) 11-16.
[x]
See http://www.ncbi.nlm.gov/BLAST/blast_databases.html
[xi]
Medini, D. and Beltrami, R. Biowulf Blast in a Solaris environment: a
preliminary test, technical annex for the present Project.
[xii]
Beltrami, R. et al., Deliverable 4.1 of the present Project.
[xiii]
Berendsen, H.J.C. et al. cited.
[xiv] Rossi, I. and Fariselli, P.
Deliverable 3.1 of the present Project.
[xv] Rossi, I. and
Fariselli, P. cited
[xvi] Altschul, S.F., et al., Basic local
alignment search tool, J. Mol. Biol. 215 (1990) 403-410
[xvii]
Geist, A. et al., PVM: Parallel Virtual Machine; A Users' Guide and
Tutorial for Networked Parallel Computing, MIT Press, Cambridge, MA,
1994.
[xviii] Message Passing Interface Forum, "MPI: A
message passing interface standard" International Journal of
Supercomputer Applications, 8 (3/4) 1994.
[xix] See
http://www-unix.mcs.anl.gov/mpi/mpich/
[xx] Rossi, I. and
Martelli, P.L. Deliverable 3.3 of the present Project.
[xxi] See
http://www.sun.com/software/hpc
[xxii] Rossi, I. and Fariselli, P.
cited.
[xxiii] Medini, D. and Beltrami, R. Biowulf Blast in a
Solaris environment: a preliminary test, technical annex for the
present Project.
[xxiv] Rossi, I. and Martelli, P.L. cited.
[xxv]
Amdahl, G.M. Validity of the single-processor approach to achieving
large scale computing capabilities. In AFIPS Conference Proceedings,
vol. 30 (Atlantic City, N.J., Apr. 18-20). AFIPS Press, Reston, Va.,
1967, pp. 483-485.
[xxvi] Medini, D. and Beltrami, R. Biowulf
Blast in a Solaris environment: a preliminary test, technical annex
for the present Project.
[xxvii] Rossi, I. and Fariselli, P.
cited.
[xxviii] Medini, D. and Beltrami, cited.